1. Statistics
2. Search DB
Here are two methods to search for data in TCMKD:
①Users can search through the search box on the homepage. Specifically, you should first select the type of data they wish to search for, then enter the data, and click on "Search" to retrieve information related to that entry.
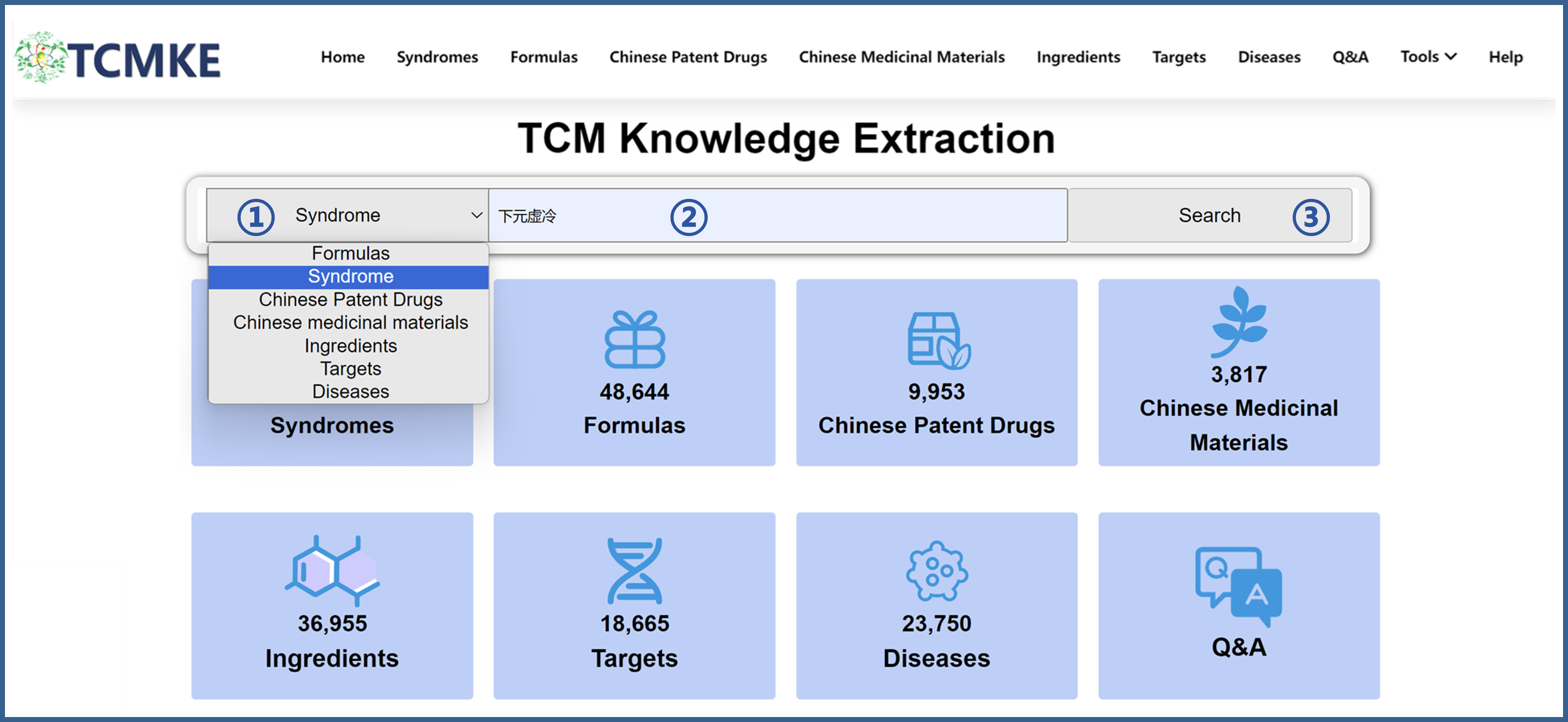
②Users can also search through the search box on the browsing interface of each data module. Taking the "Syndrome" module as an example, you can search by entering the Syndrome ID, Chinese name, pinyin name, and English name.
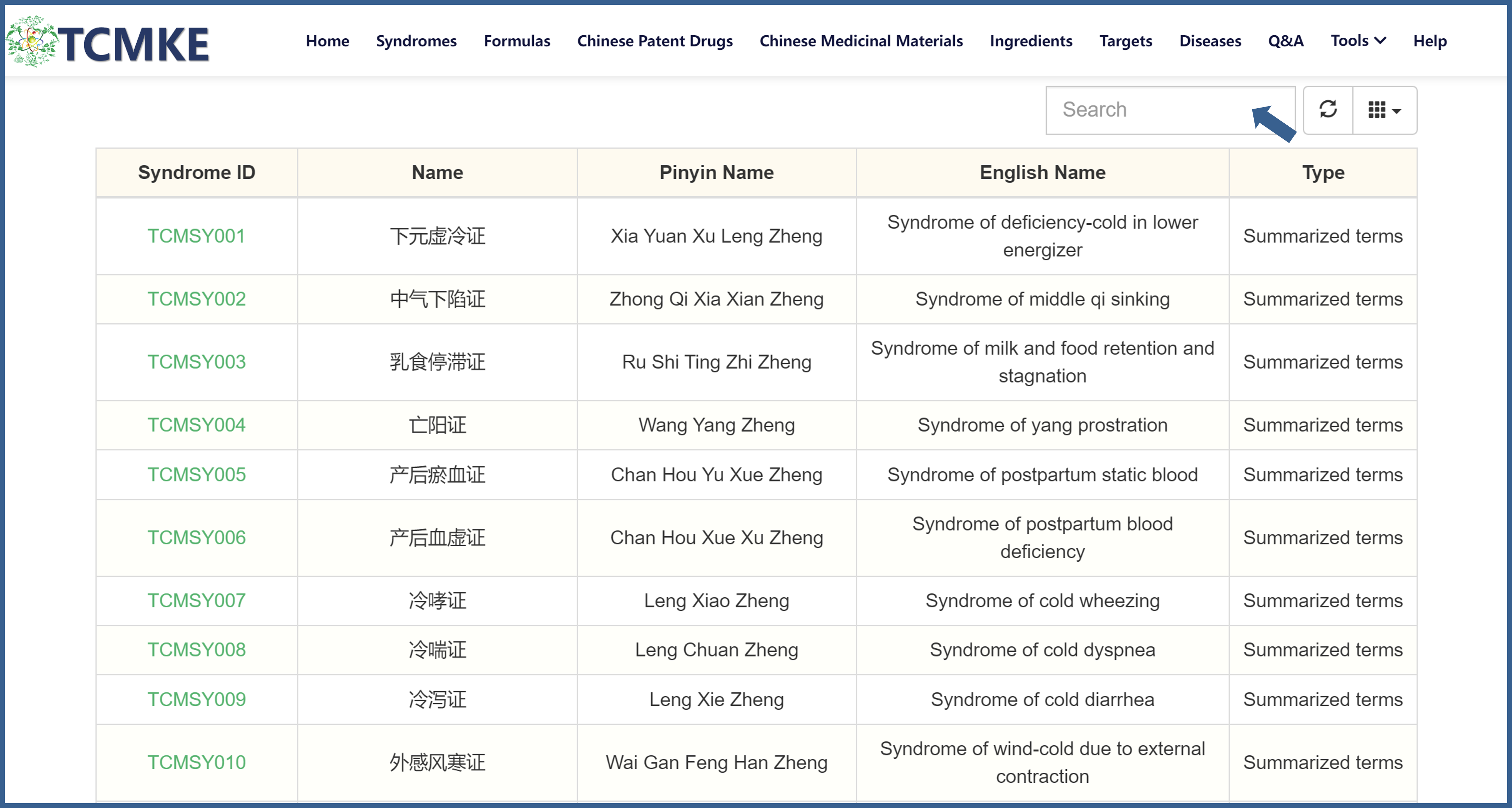
3. View the detailed page for each item
Users can access the details page by clicking on the hyperlinks in green font for a specific item from the browsing page of each data module.
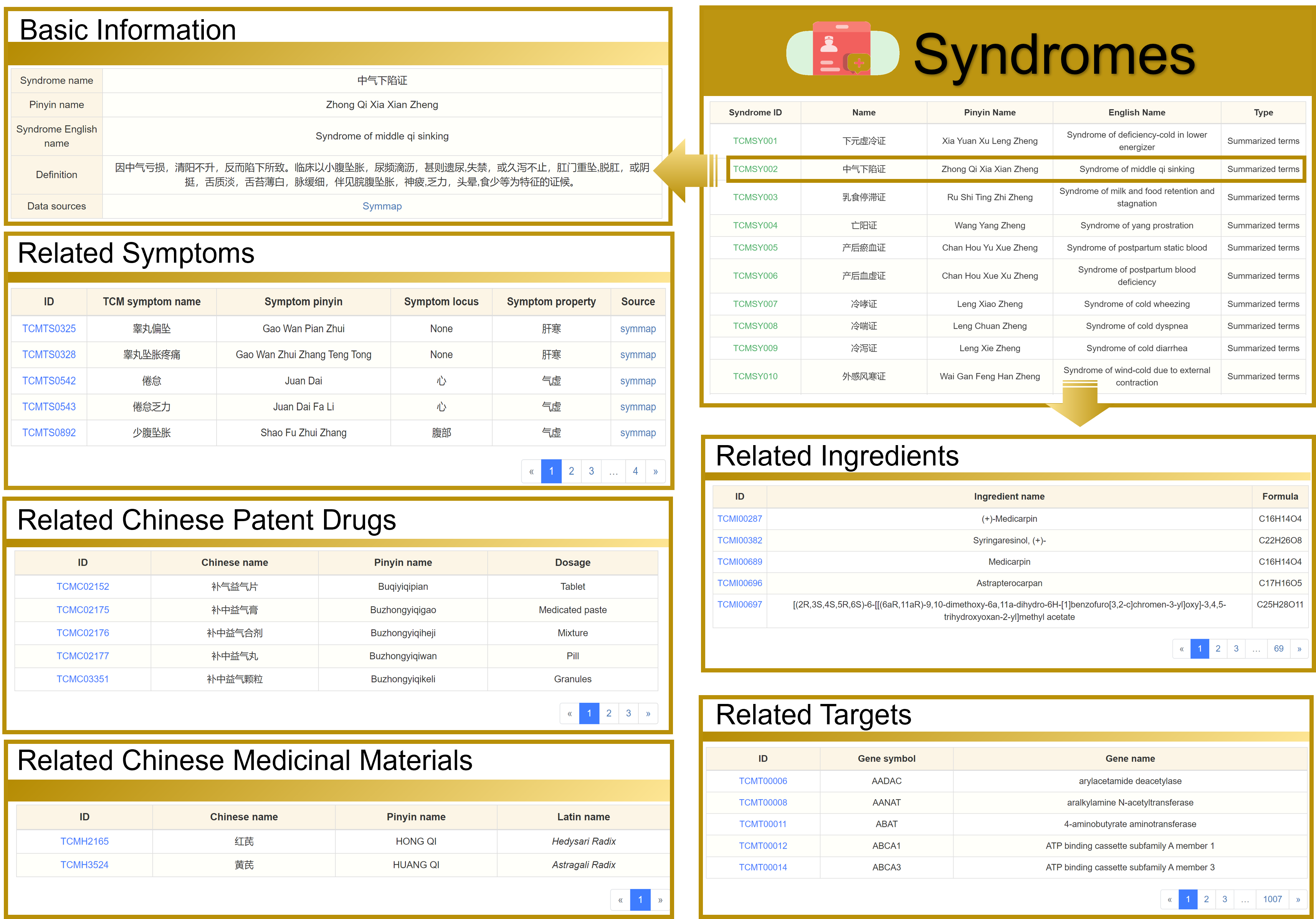
3.1 View the Syndromes detailed page
Taking "中气下陷证" as an example, the detailed page contains five sections: Basic Information, Related Symptoms, Related Chinese Patent Drugs, Related Chinese Medicinal Materials, Related Ingredients, and Related Targets. And you can jump to other interfaces by clicking on the hyperlinks on this page.
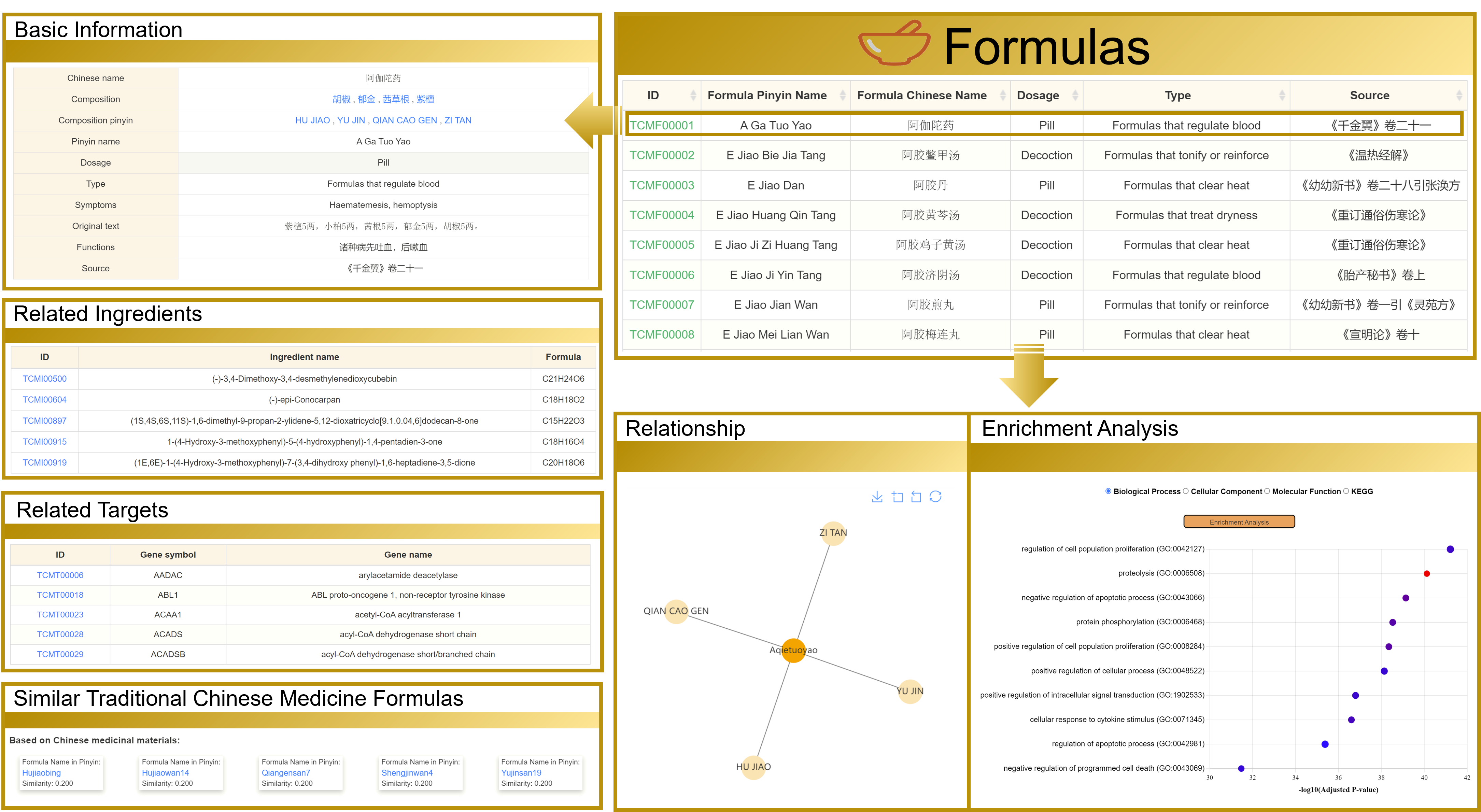
3.2 View the Formulas detailed page
Taking "阿伽陀药" as an example, the detailed page contains six sections: basic information, Related Ingredients, Related Targets, Similar Traditional Chinese Medicine Formulas, Relationship, and Enrichment Analysis. Moreover, you can navigate to other interfaces by clicking the hyperlinks present on the page.
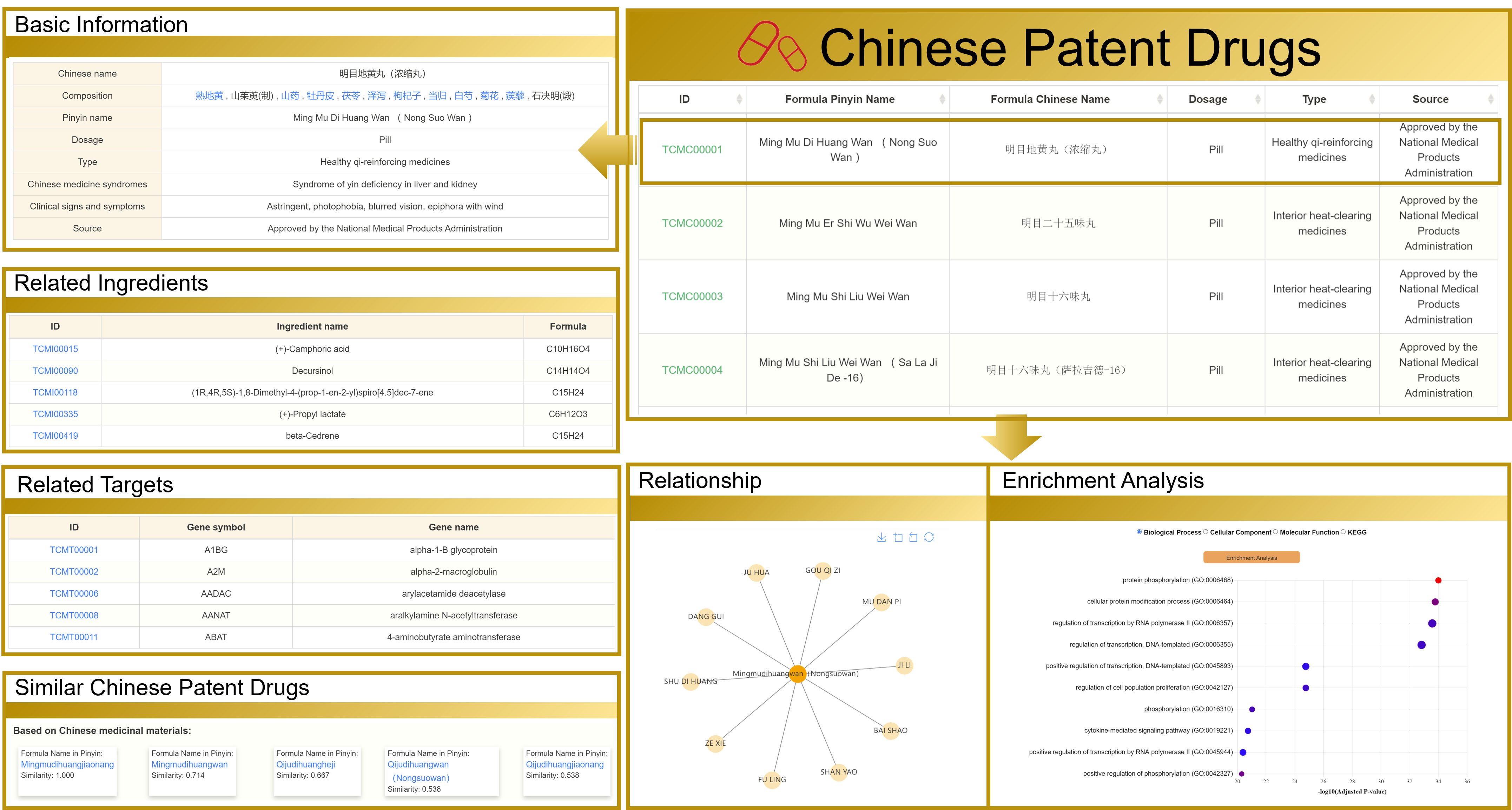
3.3 View the Chinese patent drugs detailed page
Taking "明目地黄丸(浓缩丸)" as an example, the detail page contains eight sections: Basic Information, Related Ingredients, Related Targets, Similar Traditional Chinese Medicine Formulas, KEGG analysis, GO analysis, and Relationship, and you can jump to other interfaces by clicking on the hyperlinks in blue font on this page.
3.4 View the Chinese Medicinal Materials detailed page
Taking "暴马子皮" as an example, the detail page contains eight sections: Basic Information, Related Formulas, Related Ingredients, Related Targets, KEGG analysis, GO analysis, Herb Habitat, and Relationship, and you can jump to other interfaces by clicking on the hyperlinks in blue font on this page.

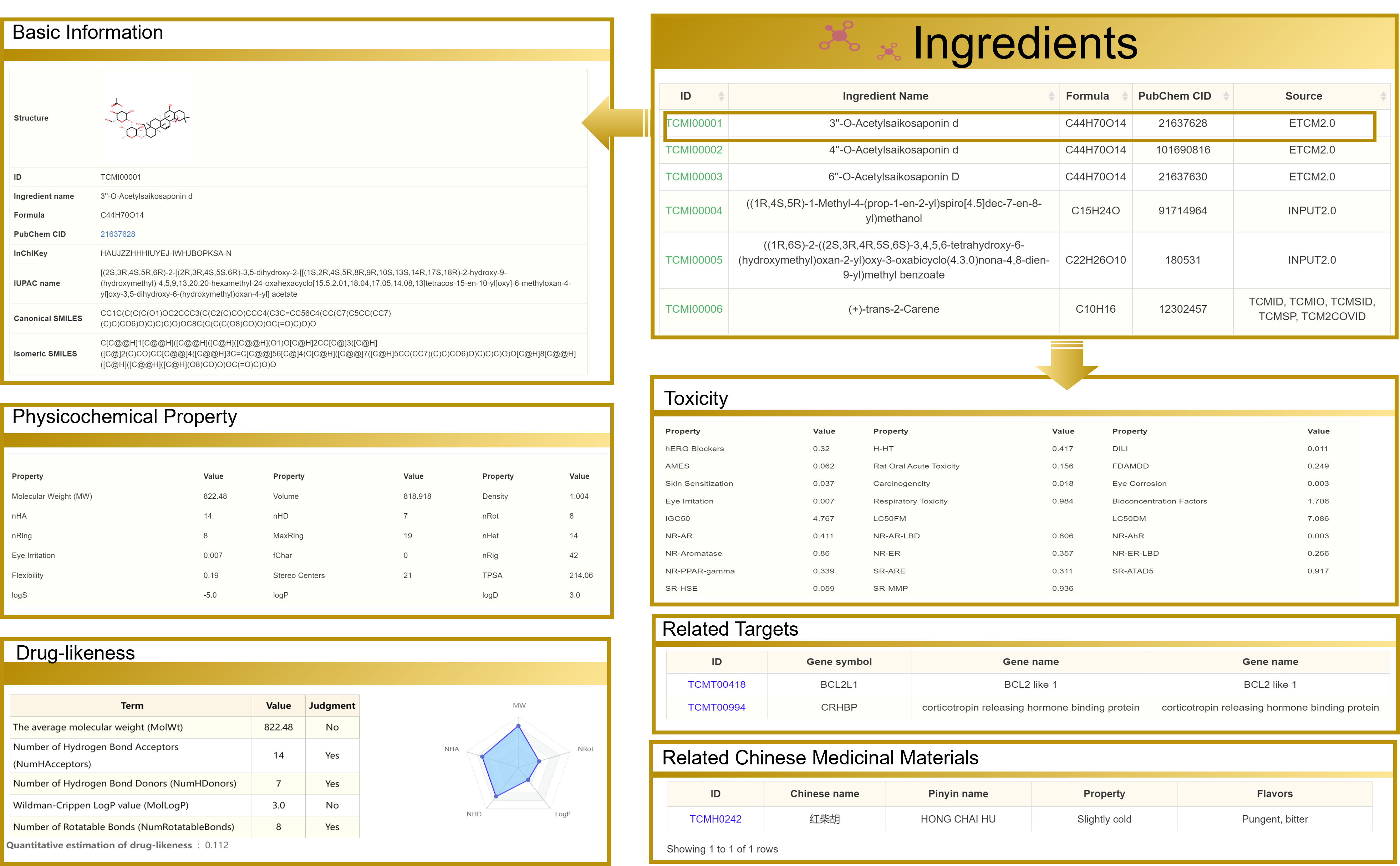
3.5 View the Ingredients detailed page
Taking "3''-O-Acetylsaikosaponin d" as an example, the detailed page contains six sections: Basic Information, Drug-likeness, Toxicity, Physicochemical Property, Related Targets, Relationship, and Related Chinese Medicinal Materials. Moreover, you can navigate to other interfaces by clicking the hyperlinks present on the page.
3.6 View the Targets detailed page
Taking "NAT2" as an example, the detailed page contains three sections: Basic Information, Related Chinese Medicinal Materials, and Related Targets. Moreover, you can navigate to other interfaces by clicking the hyperlinks present on the page.

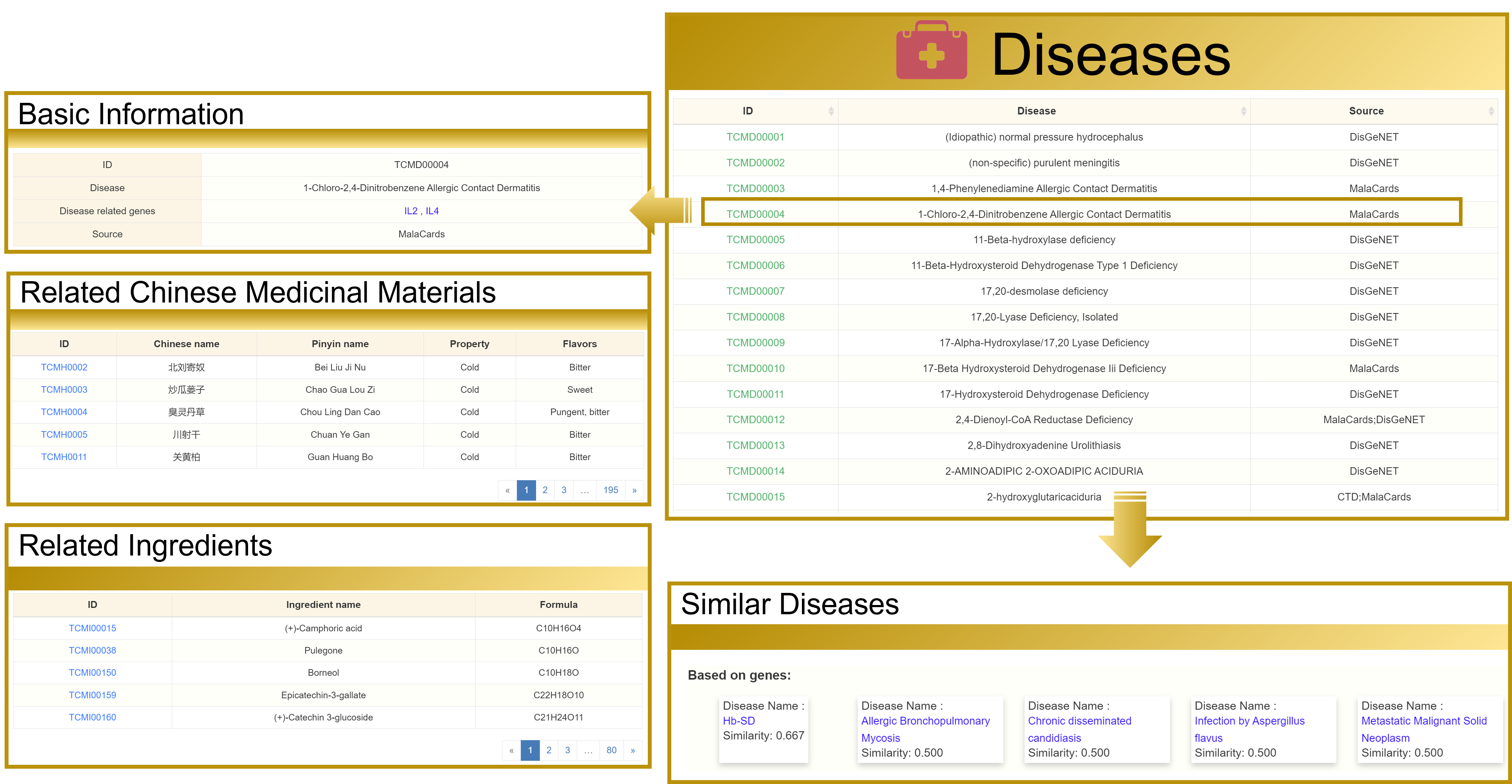
3.7 View the Diseases detailed page
Taking " 1-Chloro-2,4-Dinitrobenzene Allergic Contact Dermatitis" as an example, the detailed page contains four sections: Basic Information, Related Chinese Medicinal Materials, Related Ingredients, and Similar Disease, you can jump to other interfaces by clicking on the hyperlinks in blue font on this page.
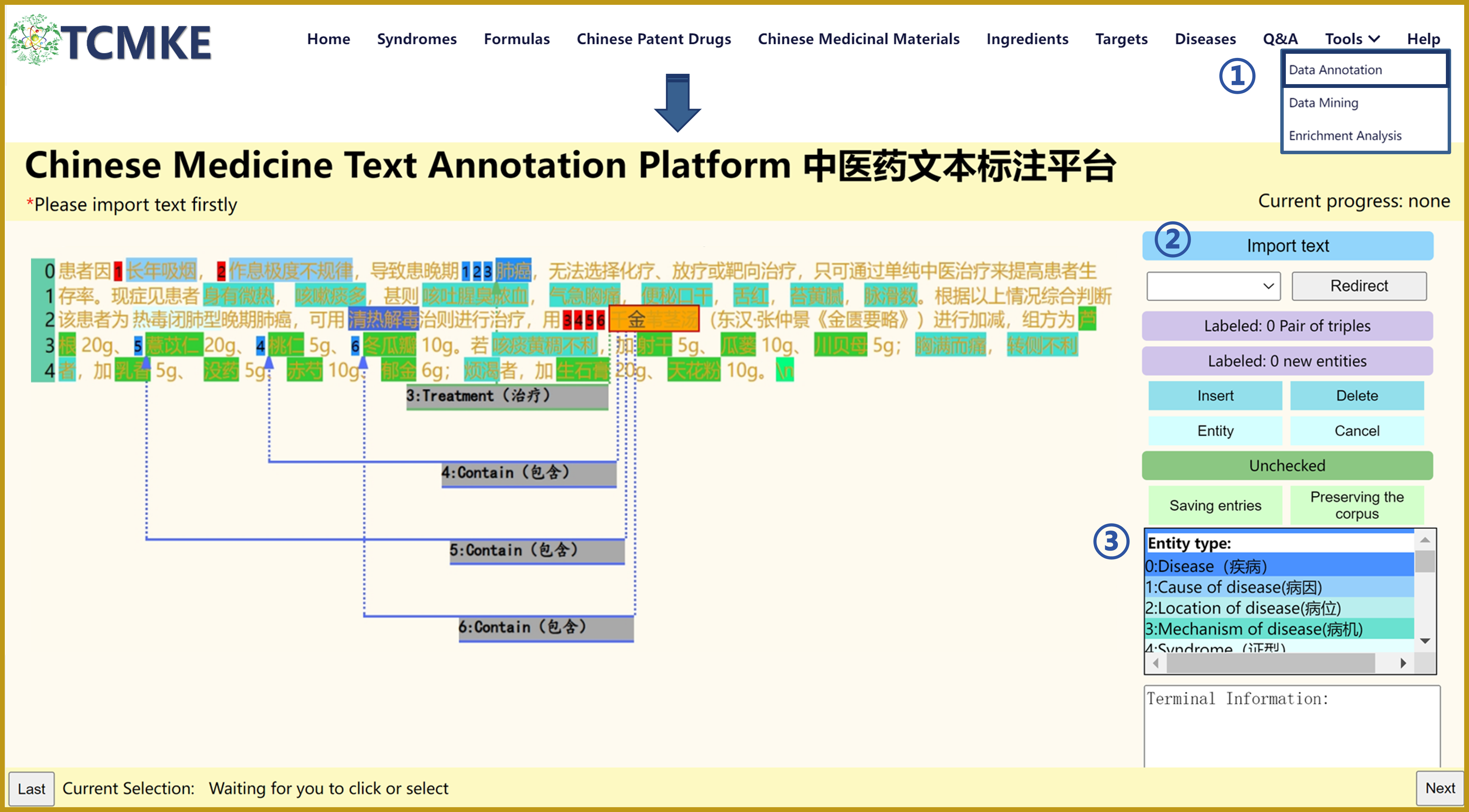
4. Use of data annotation tool
①Click "Tools" and select "Data Annotation" to get "Chinese Medicine Text Annotation Platform". On the right side is the toolbar where users can annotate the data.
②Click "Import text" to select the document to be uploaded for annotation.
③Annotate the text. According to the different needs of users, the toolbar provides a series of entity types including: "Disease, " "Cause of disease, " "Location of disease," "Mechanism of disease," "Syndrome," "Symptom," "Method of treatment," "Therapeutic principle," "Formula," "Chinese patent drug," "Type," "Herb," "Property," "Flavor," "Meridian entry," "Preparation Methods," "Pharmaceutical ingredients, and Acupuncture point. The types of relationships provided for entities are Belong to, Property, Flavor, Meridian entry, Type, Treatment, Use, Therapeutic principle, Method of treatment, Contain, Mechanism of disease, Location of disease, and Cause of disease.
Additionally, click on the appropriate button for the entity "Insert," "Delete," "Entity," and "Cancel." The platforms can carry out the corresponding reactions. Users can also save part of the labeled data by clicking on "Saving entries," and "Preserving the corpus."
5. Use of data mining tool
Click "Tools" and select "Data Mining" to get the "TCM Data Mining". Here are four analysis tools.
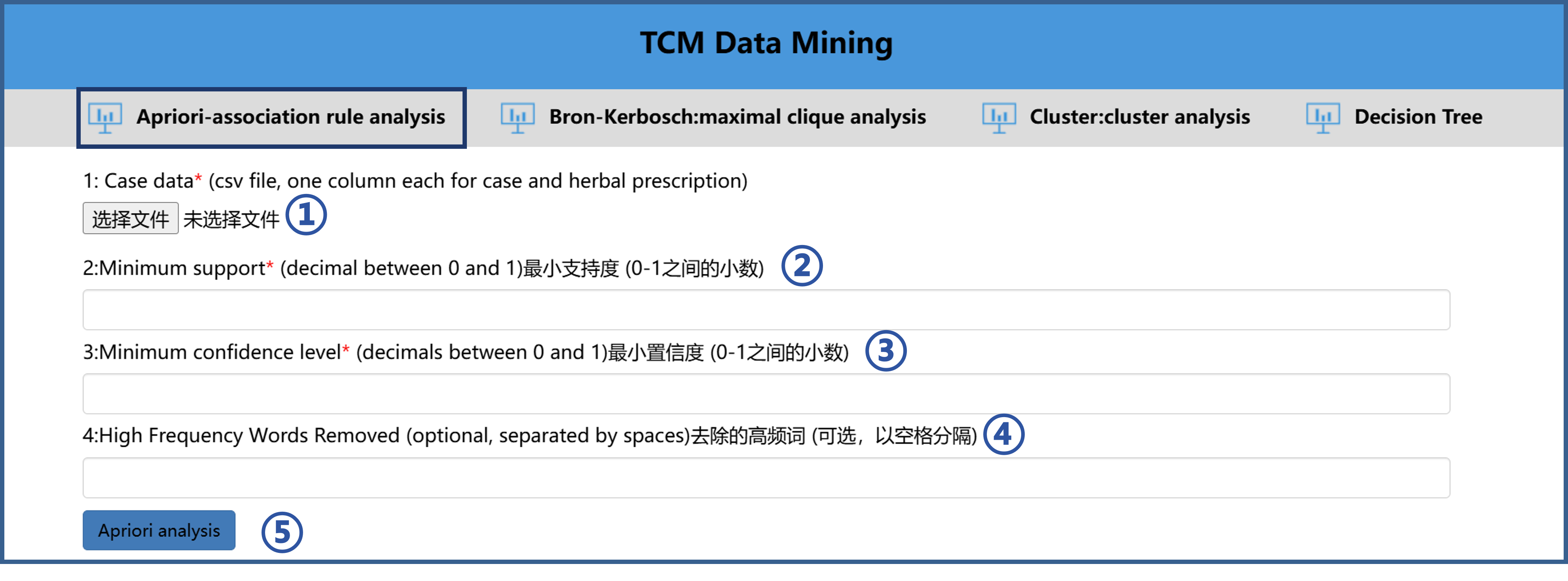
5.1 Use of Apriori-association rule analysis
The operation steps of Apriori-association rule analysis tool are as follows.
①Import the file to be analyzed according to the format of the sample file.
②Set the Minimum support.
③Set the Minimum confidence level.
④If you need to remove high-frequency words, then remove the high-frequency words (not necessary).
⑤ Click “Apriori analysis” button to analyze.
5.2 Use of maximal clique analysis
The operation steps of maximal clique analysis tool are as follows.① Import the file to be analyzed according to the format of the sample file.
② Set the minimum co-present number.
③ Enter the CBWN.
④ Enter the SBCα.
⑤ Enter the minimum number of co-occurrences of the plotted graph.
⑥ Click “BK analysis” button to analyze.
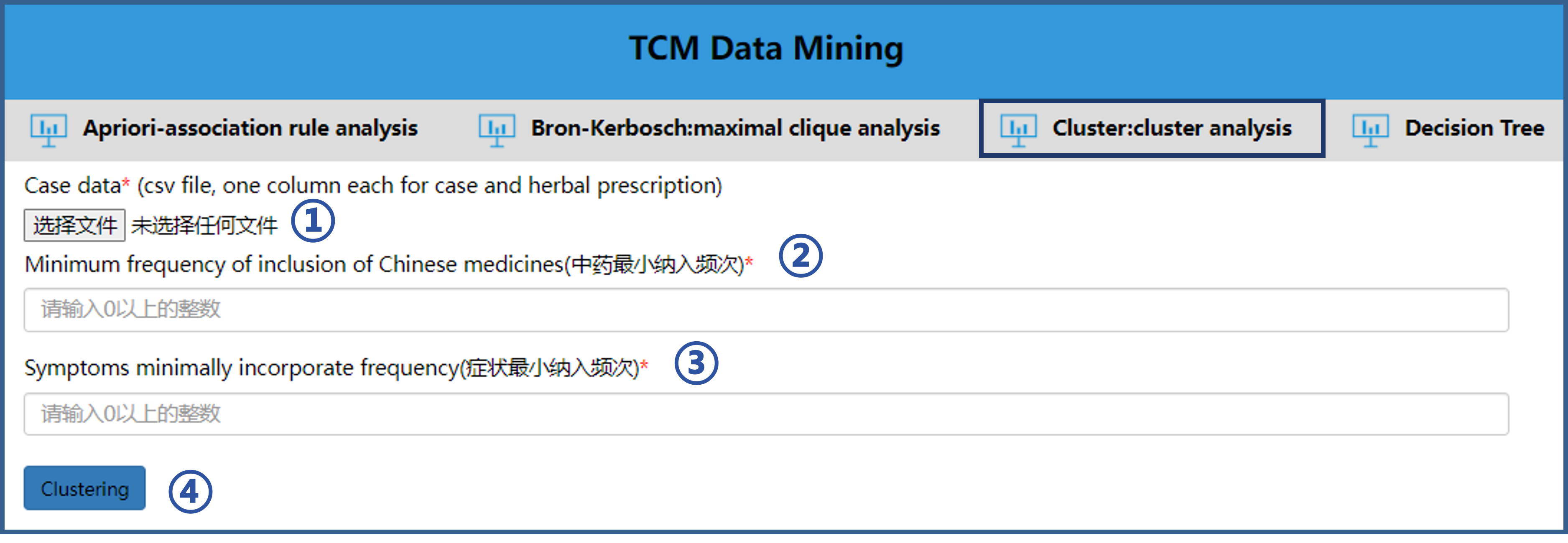
5.3 Use of cluster analysis
The operation steps of cluster analysis tool are as follows.① Import the file to be analyzed according to the format of the sample file.
② Enter the minimum frequency of inclusion of herbs.
③ Enter the syndromes minimally incorporate frequency.
④ Click “Clustering” button to analyze.
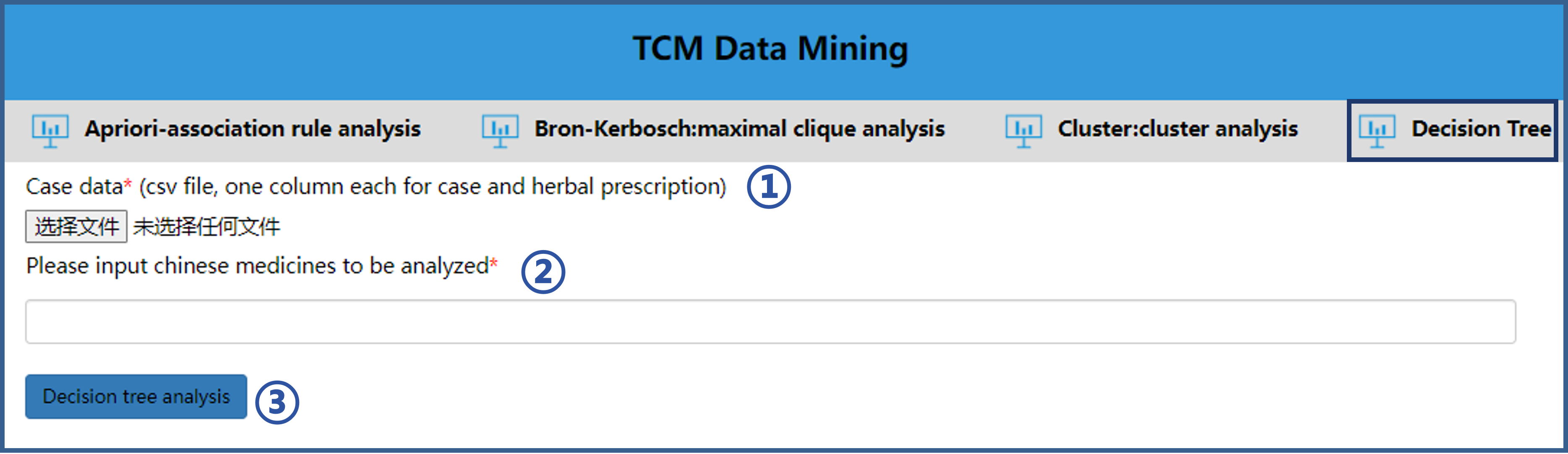
5.4 Use of Decision Tree analysis
The operation steps of Decision Tree analysis tool are as follows.① Import the file to be analyzed according to the format of the sample file.
② Input the herbs to be analyzed.
③ Click “Decision tree analysis” button to analyze.
6. Use of enrichment analysis tool
Users can perform enrichment analysis by following the steps below.
Here are two methods to perform enrichment analysis:
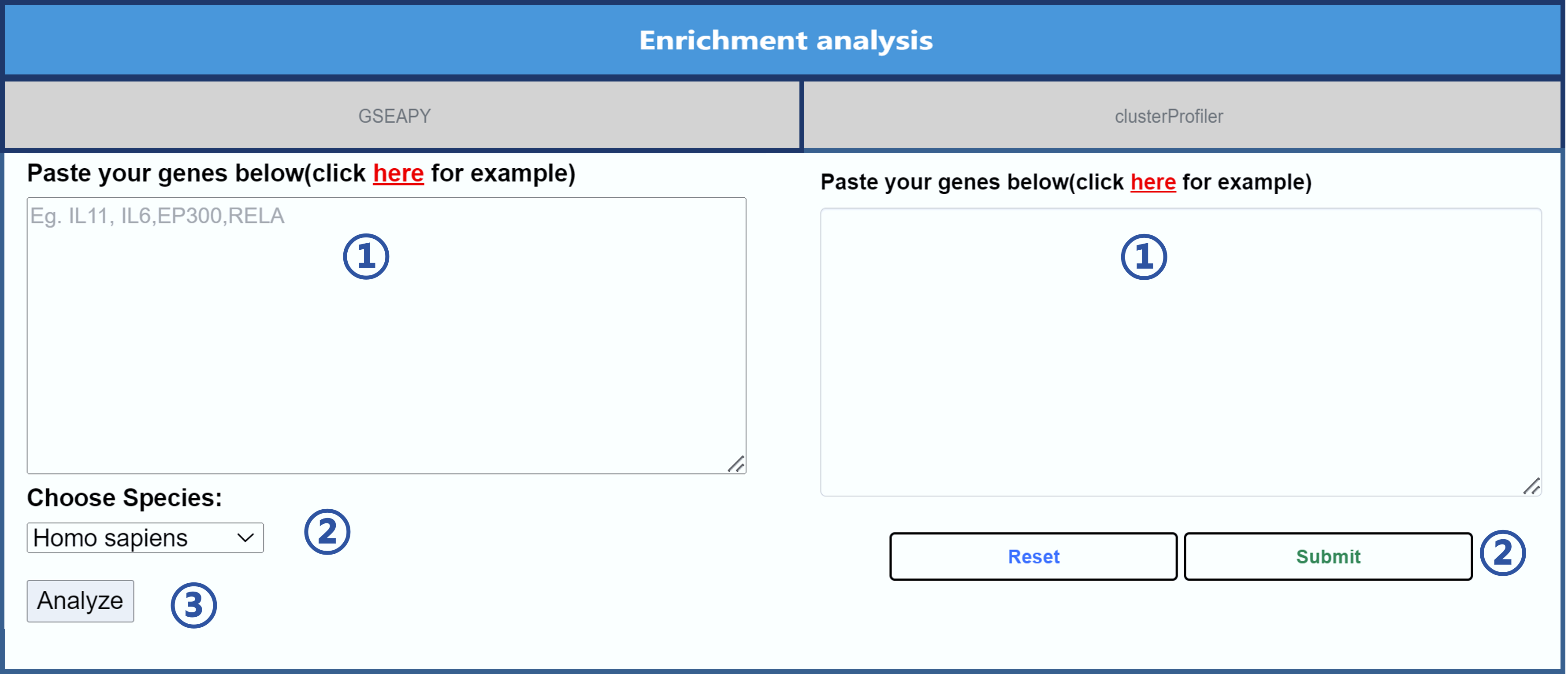
For the GSEAPY part, users can perform enrichment analysis by following the steps below.
① Enter the gene symbol to be analyzed.
② Select the species, including Homo sapiens, Mus musculus, and Rattus norvegicus.
③ Click "Analyze".
For the clusterProfiler part, users can perform enrichment analysis by following the steps below.
① Enter the gene symbol to be analyzed.
② Click "Submit".
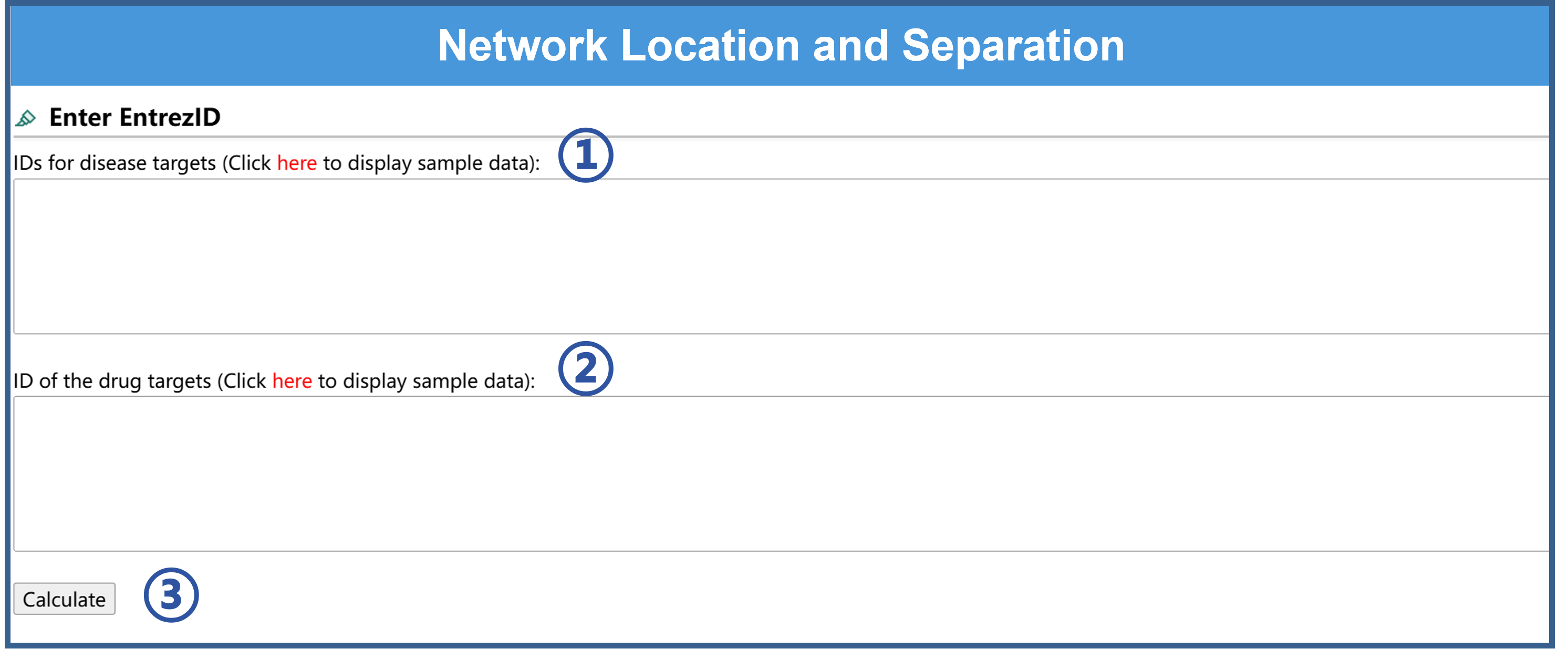
7.Use of the Network Location and Separation analysis tool
Users only need three steps to perform the Network Location and Separation analysis.
①Input one group's gene IDs of the two groups of data.
②Input the other group's gene IDs of the two groups of data.
③Click “Calculate”, then the network localization and separation analysis can be calculated and the corresponding results can be obtained.
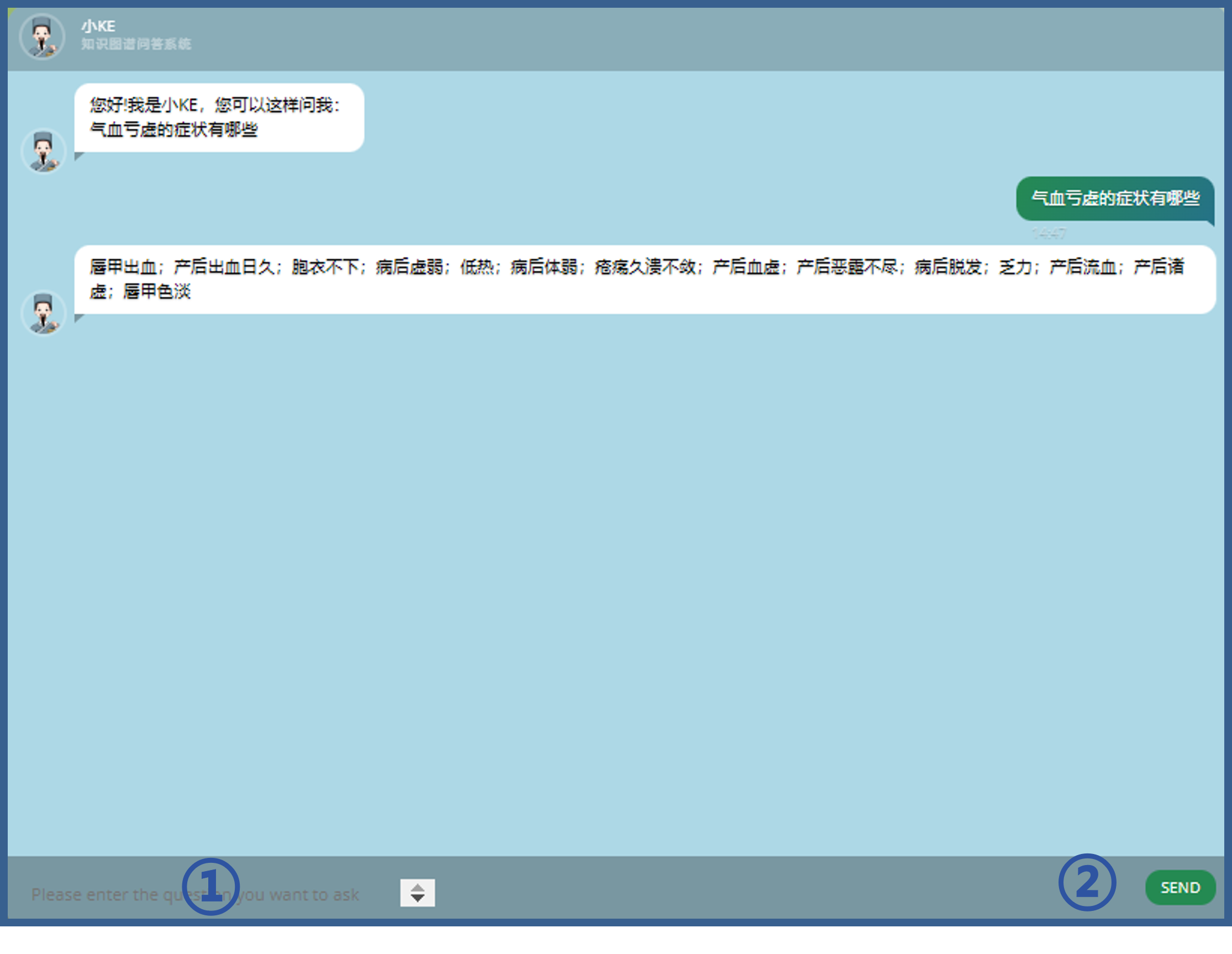
8.Use of the Q&A system
Users can make queries using the Q&A system in just two steps as follows.
① Enter what they want to query in the input box.
② Click the "Send" button will trigger the query process.
The system will answer based on the knowledge graph relationships constructed in TCMKD.
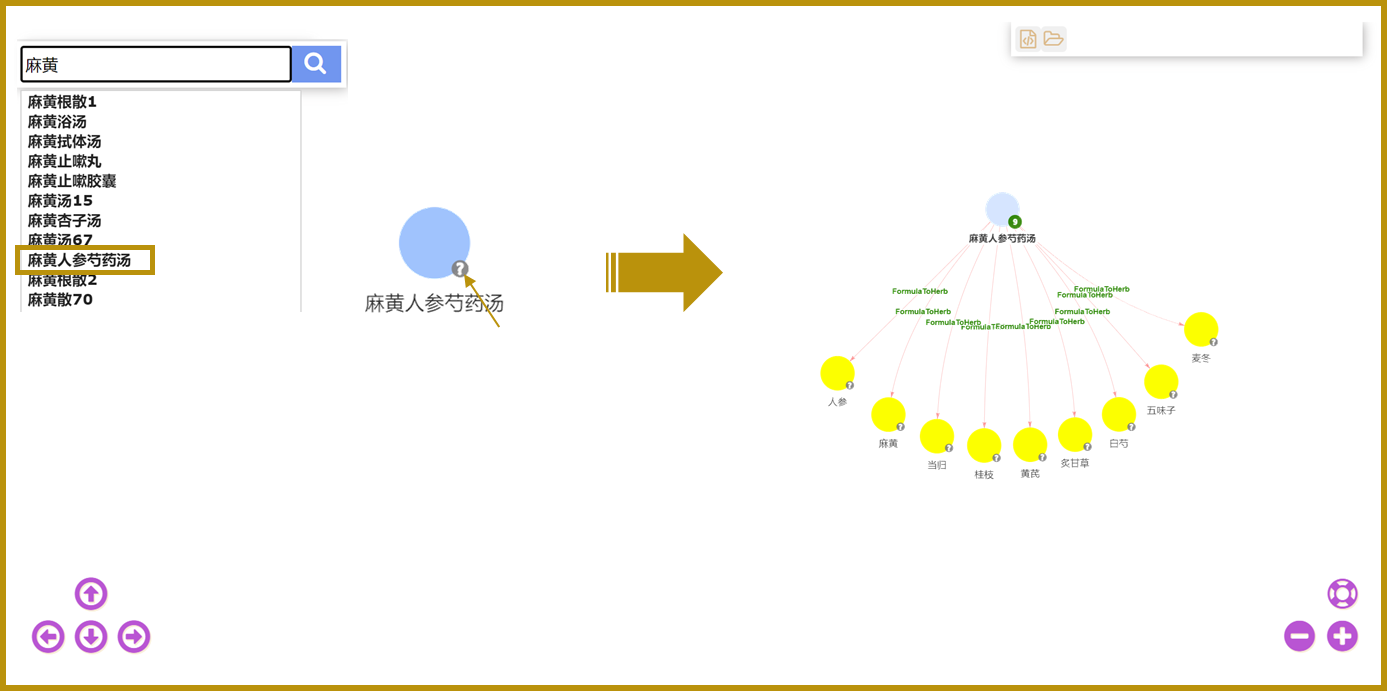
9.Use of the knowledge graph query
Knowledge Graph is a knowledge graph of TCM built on Neo4j platform. The page utilizes the Neo4j database to store information about entities, entity attributes, and relationships between entities in a graph structure. It is embedded in the website for users to browse and search. Our query system supports fuzzy query function, which means that users can retrieve relevant TCM knowledge by inputting partial information of keywords.
For example, users only need to input " 麻黄" to find the formula "麻黄人参芍药汤" in the search results.
Users can click on the node in the graph and get the detailed information of the entity in the upper left corner. Double click on the node to get the related compositions of the formula. The query results of the whole knowledge graph are presented graphically, which visually demonstrates the network structure among TCM knowledge.
11.Contact
If you use TCMKD for research, please cite the following paper: "TCMKD: From Ancient Wisdom to Modern Insights—A Comprehensive Platform for Traditional Chinese Medicine Knowledge Discovery". If you have any comments, corrections or questions please contact us.
Information about us
Email: greatchen@cdutcm.edu.cn
Email: xiaowenke@cdutcm.edu.cn
More information about us, please check https://lab.rjmart.cn/10016/ChenLab
10.Download
| Data type | Description | No. | Download |
|---|---|---|---|
| Syndromes info | The detailed information of each syndrome | 262 | Download |
| Formulas info | The detailed information of each formula | 48,644 | Download |
| Chinese Patent Drugs info | The detailed information of each Chinese Patent Drug | 9,953 | Download |
| Chinese Medicinal Materials info | The detailed information of each Chinese Medicinal Material | 3,817 | Download |
| Ingredients info | The detailed information of each ingredient | 36,403 | Download |
| Targets info | The detailed information of each target gene | 18,655 | Download |
| Diseases info | The detailed information of each disease | 23,750 | Download |
