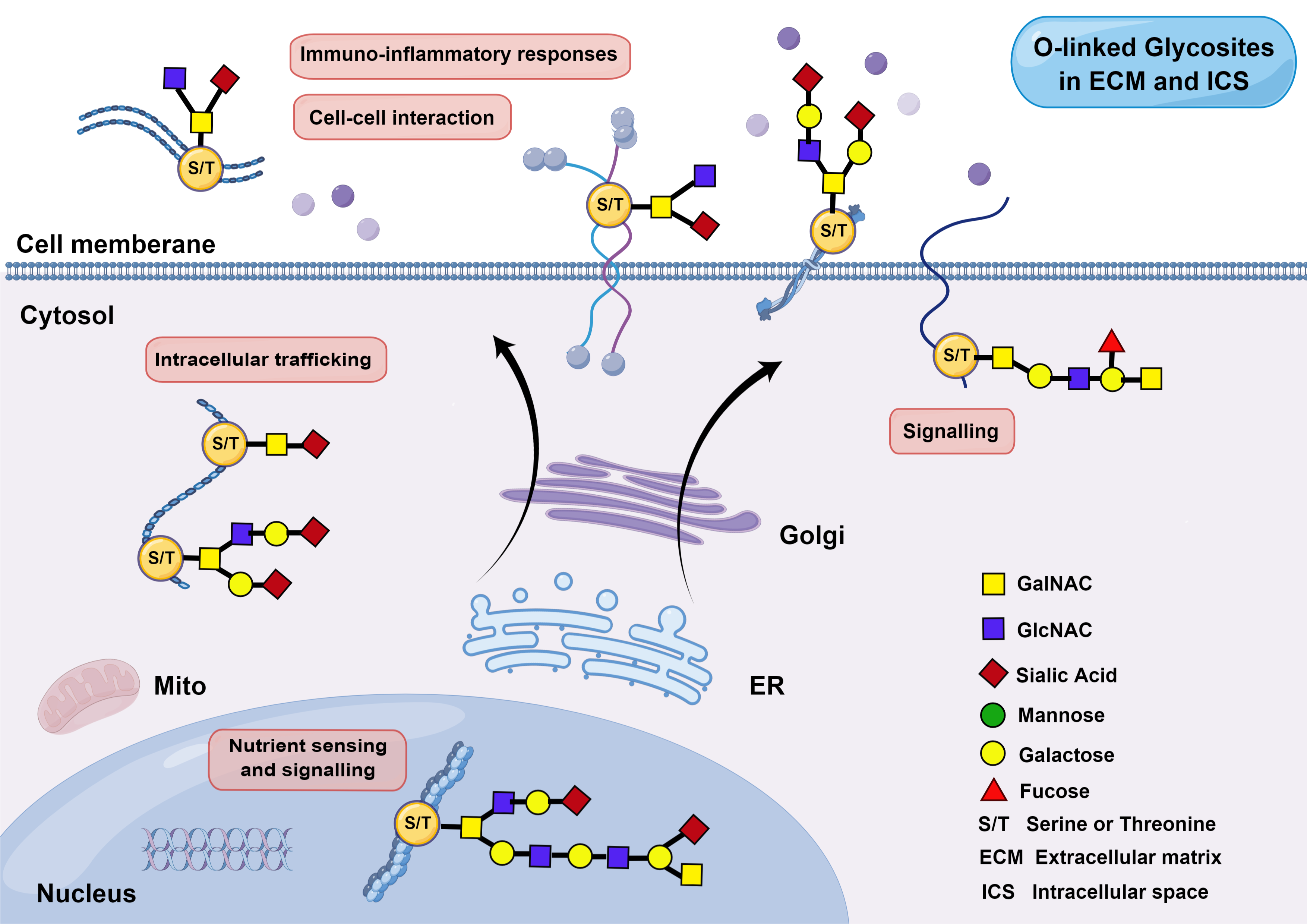
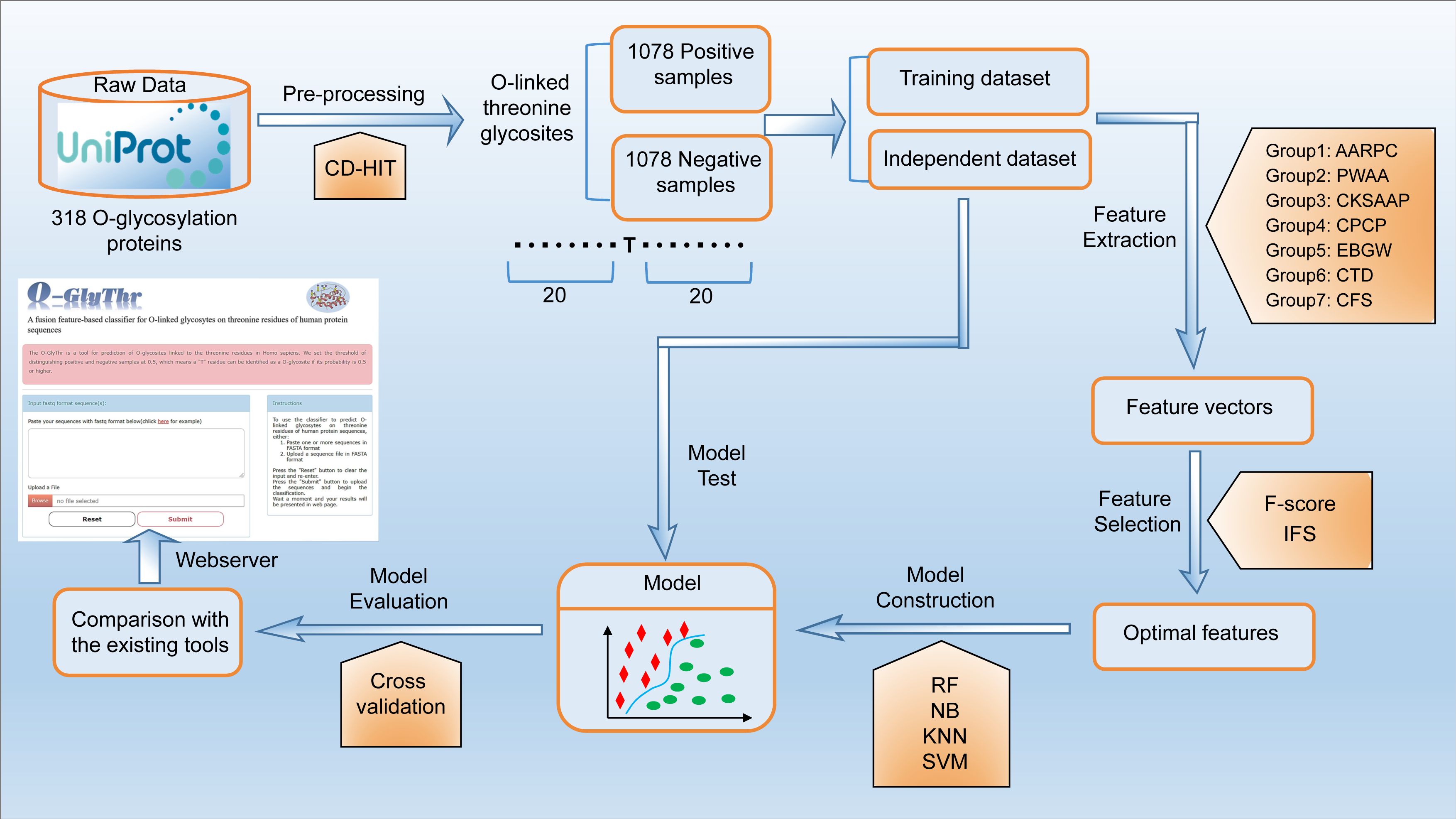
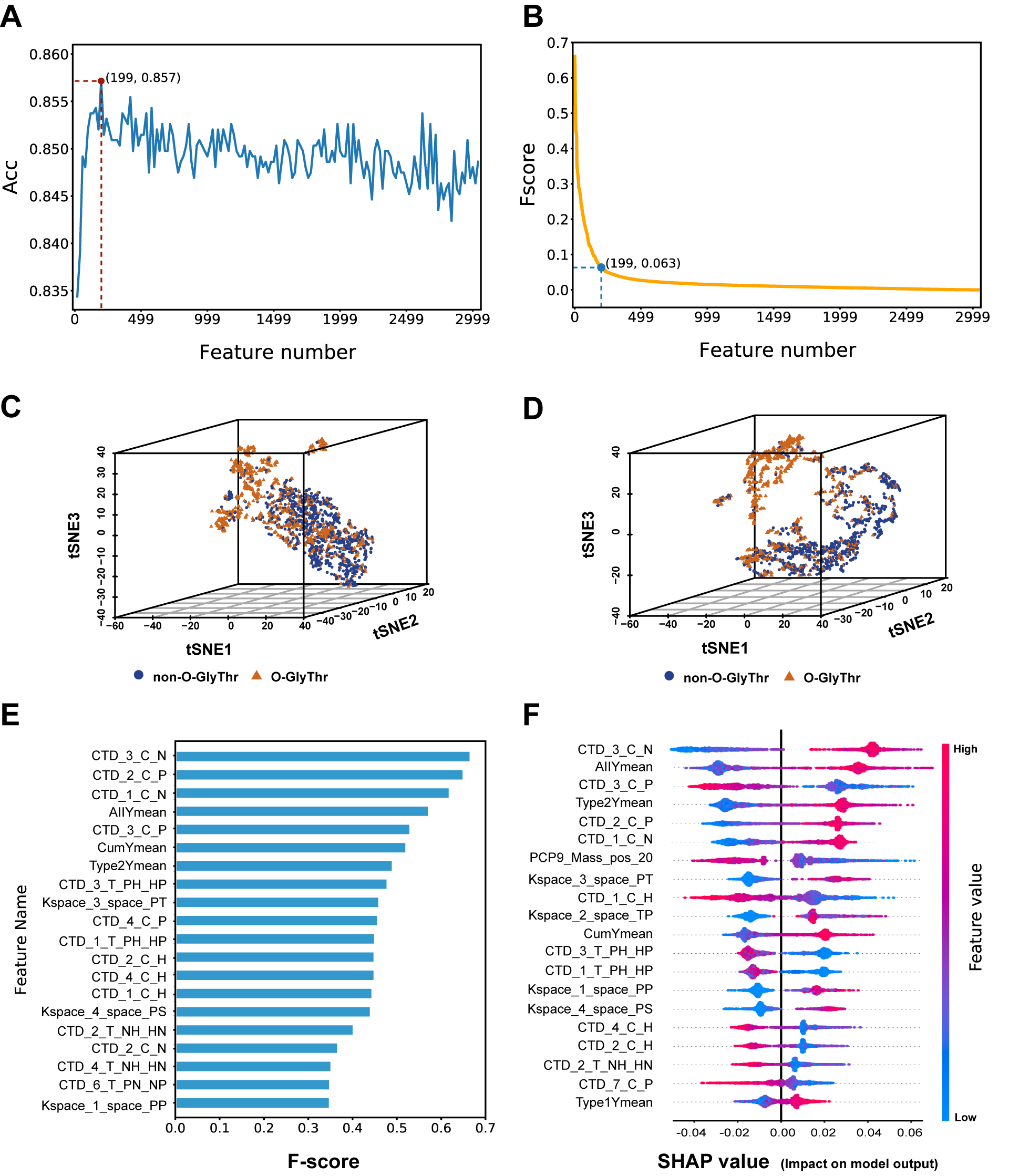
Introduction to O-GlyThr
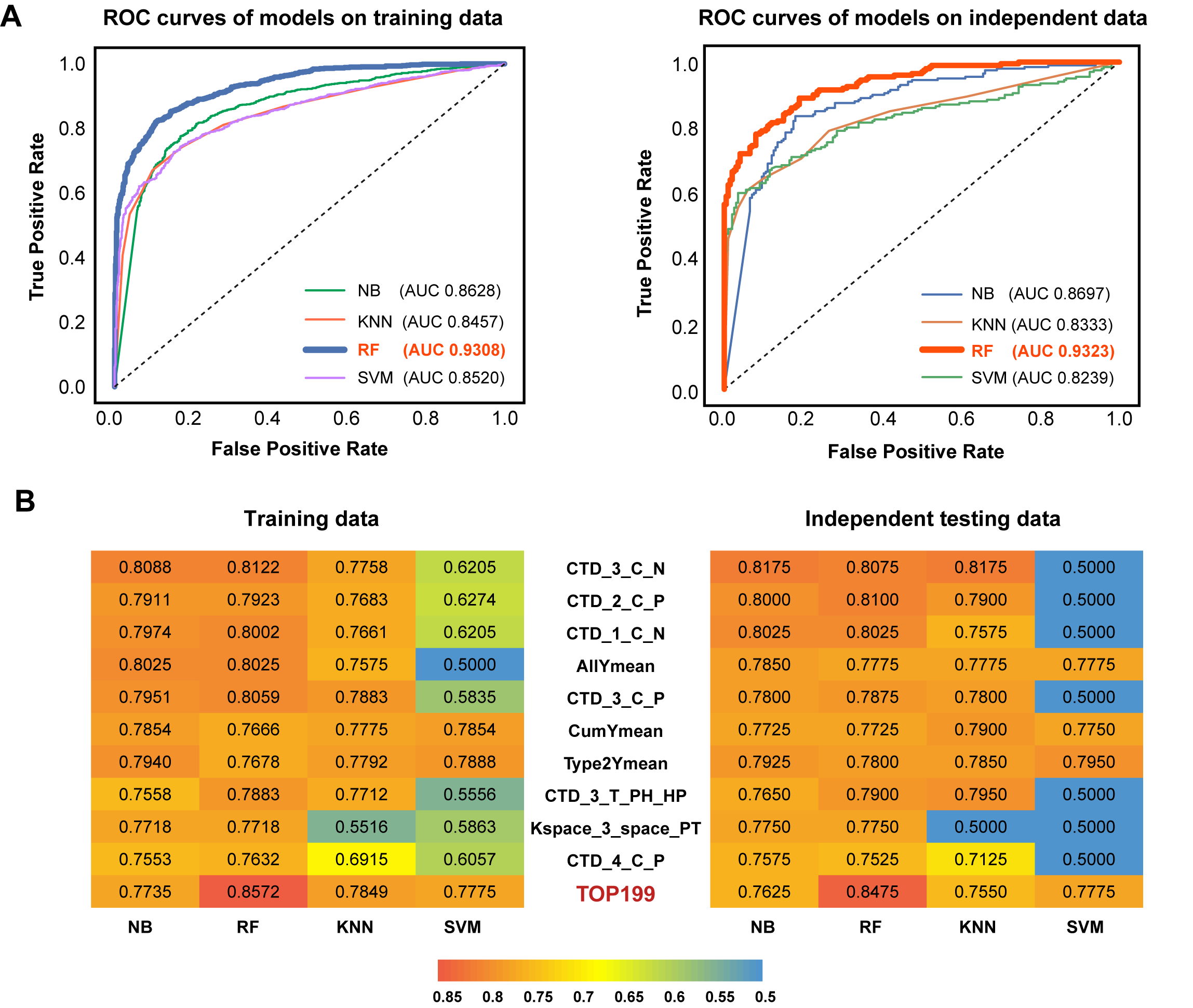
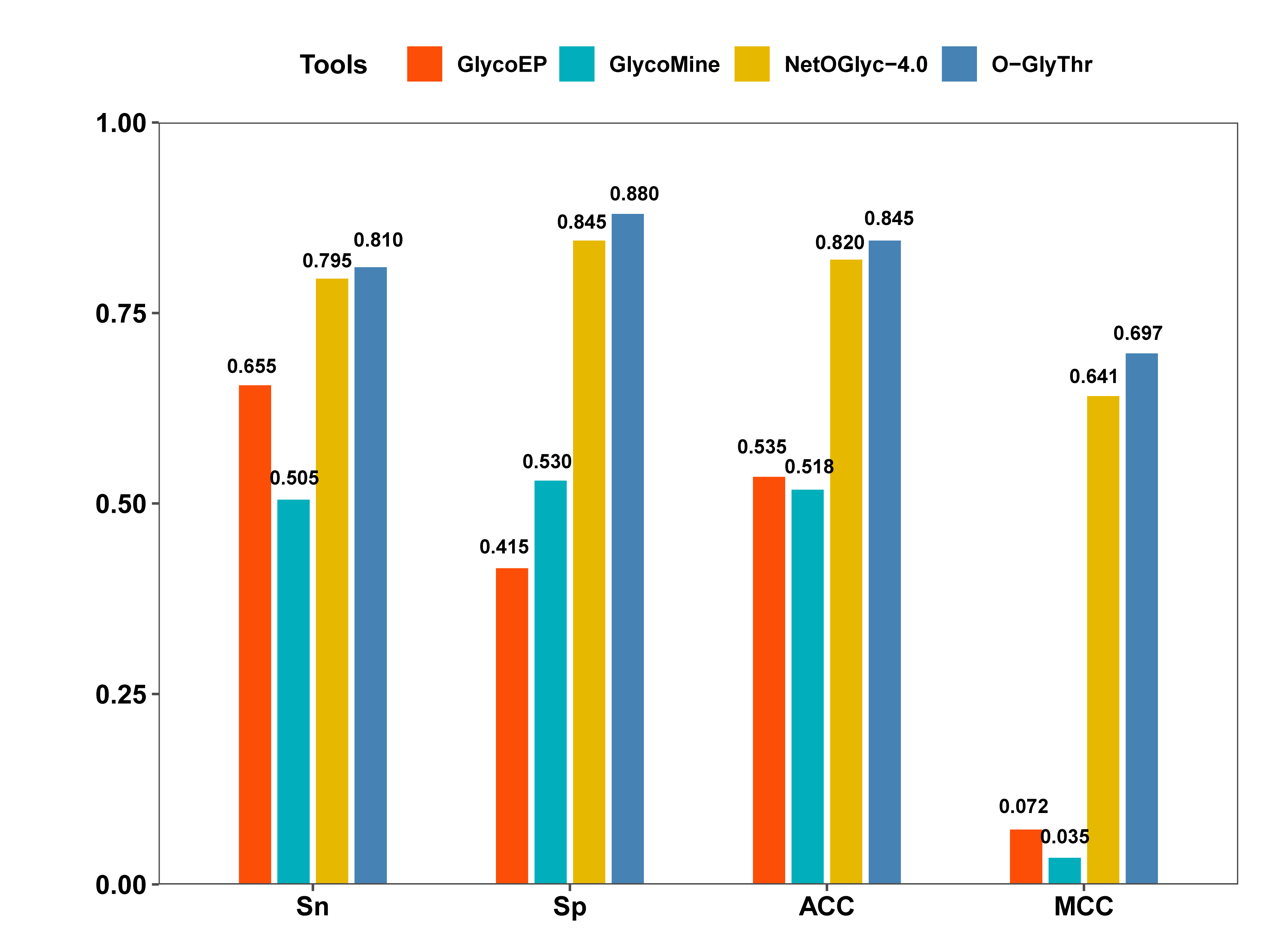
A prediction model based on feature fusion was constructed for O-glycosites linked to the threonine residues in Homo sapiens. High-quality human protein data with O-linked threonine glycosites were collected and sorted out. Seven feature coding methods were fused to represent the sample sequence. Random forest, outweighing three other machine learning algorithms, was selected as the final classifier to construct the classification model. The validation on the independent test dataset demonstrated the superiority of O-GlyThr to the other available online predictors..