The INPUT provides the Network Pharmacology, Enrichment Analysis and Data Query options. In order to make sure INPUT is convenient to be used,the help page is prepared.
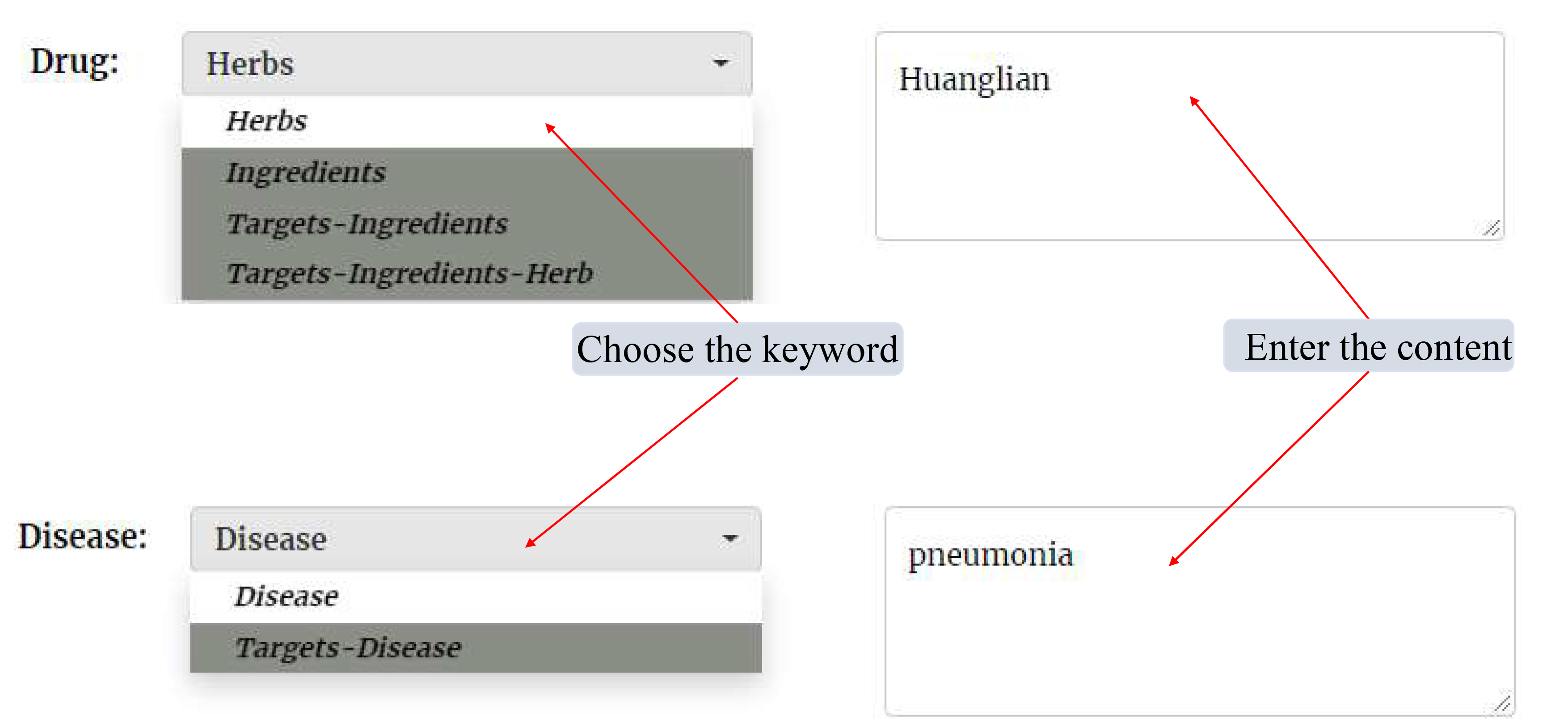
For the Network Pharmacology page, users need to enter the content in the "Drug" module and the "Disease" module separately
(1) In the "Drug" module, if users choose the "Herbs" as the keyword, users need to enter the name of Herb (such as 黄连 or Huanglian); if users choose the Ingredients as the keyword, users need to enter the name or inchikey or pubchem_cid of Ingredients (such as berberine or YBHILYKTIRIUTE-UHFFFAOYSA-N or 2353); if users choose the "Targets-Ingredients" as the keyword, users need to enter the relationship between targets and ingredients (such as IL6//berberine); if users choose the Targets-Ingredients-Herb as the keyword, users need to enter the relationship between targets, ingredients and herbs (such as IL6//berberine//Huanglian).
(2) In the drop-down menu of the "Disease" module, if users choose the "Disease" as the keyword, users need to enter the name of Disease (such as pneumonia); if users choose the "Targets-Disease" as the keyword, users need to enter the relationship between targets and Disease (such as IL6//pneumonia). Then, clicking the submit button, the results will be shown in a new page.
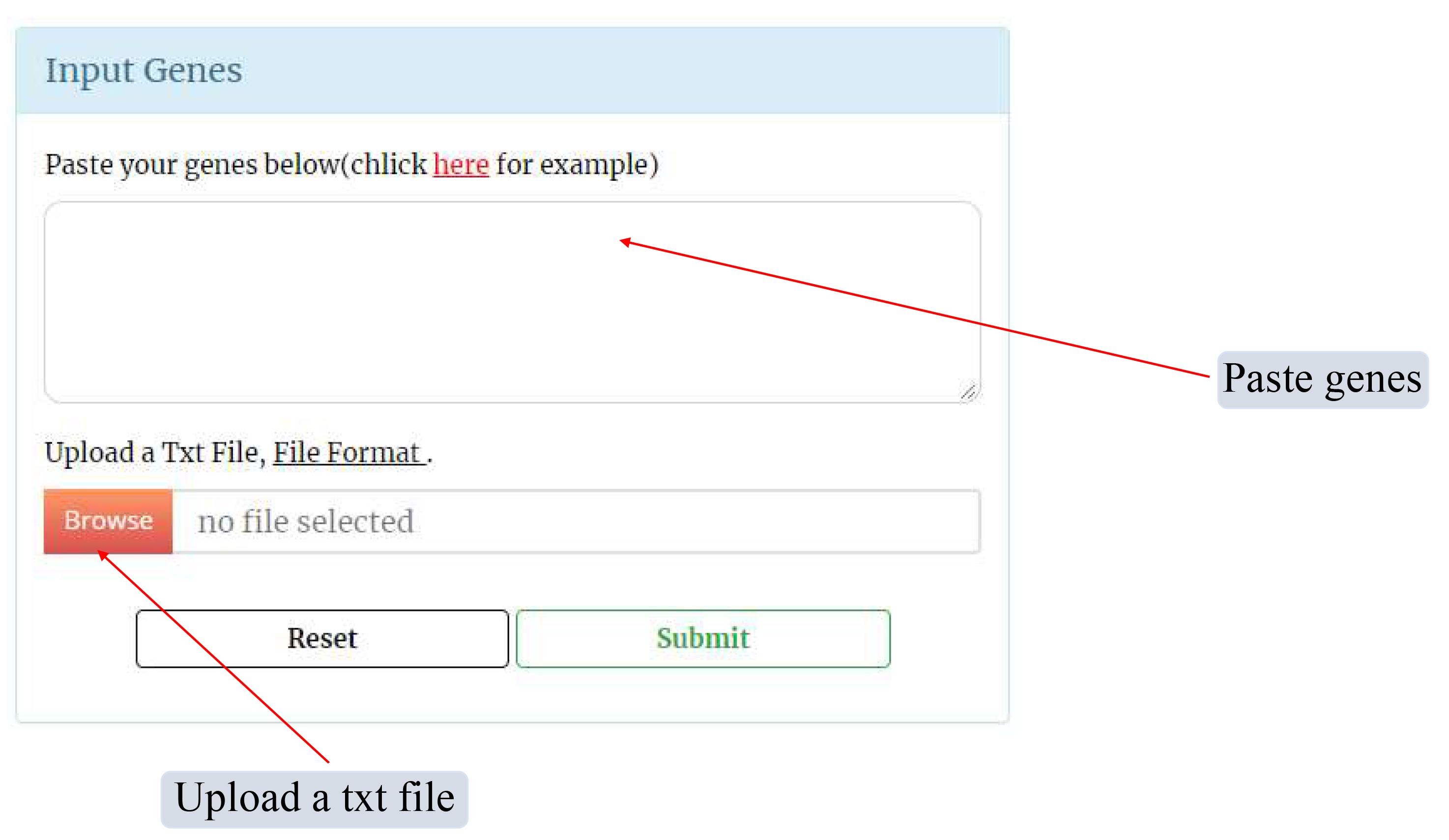
In the Enrichment Analysis page, users can paste genes or upload a text file to perform Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis.
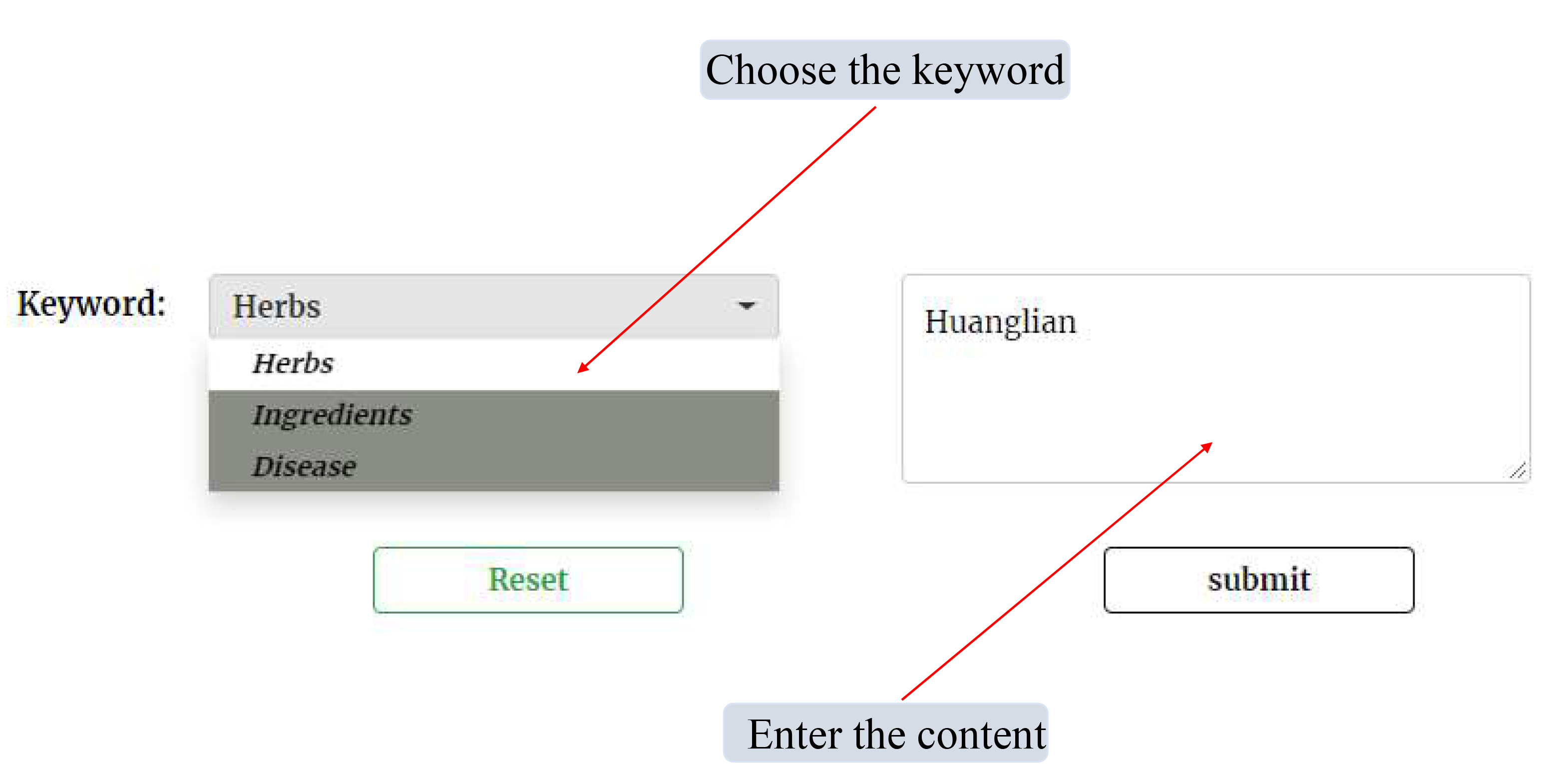
In this page, the information related to Herb, Ingredient and Disease were provided. Users can get Herb-related information by choosing the "Herbs" as the keyword, and enter the name of Herb (such as 黄连 or Huanglian); Users can get Disease-related information by choosing the "Disease" as the keyword, and enter the name of Disease (such as pneumonia); Users can also get Ingredient-related information by choosing the Ingredients as the keyword, and enter the name or inchikey or pubchem_cid of Ingredients (such as berberine or YBHILYKTIRIUTE-UHFFFAOYSA-N or 2353)
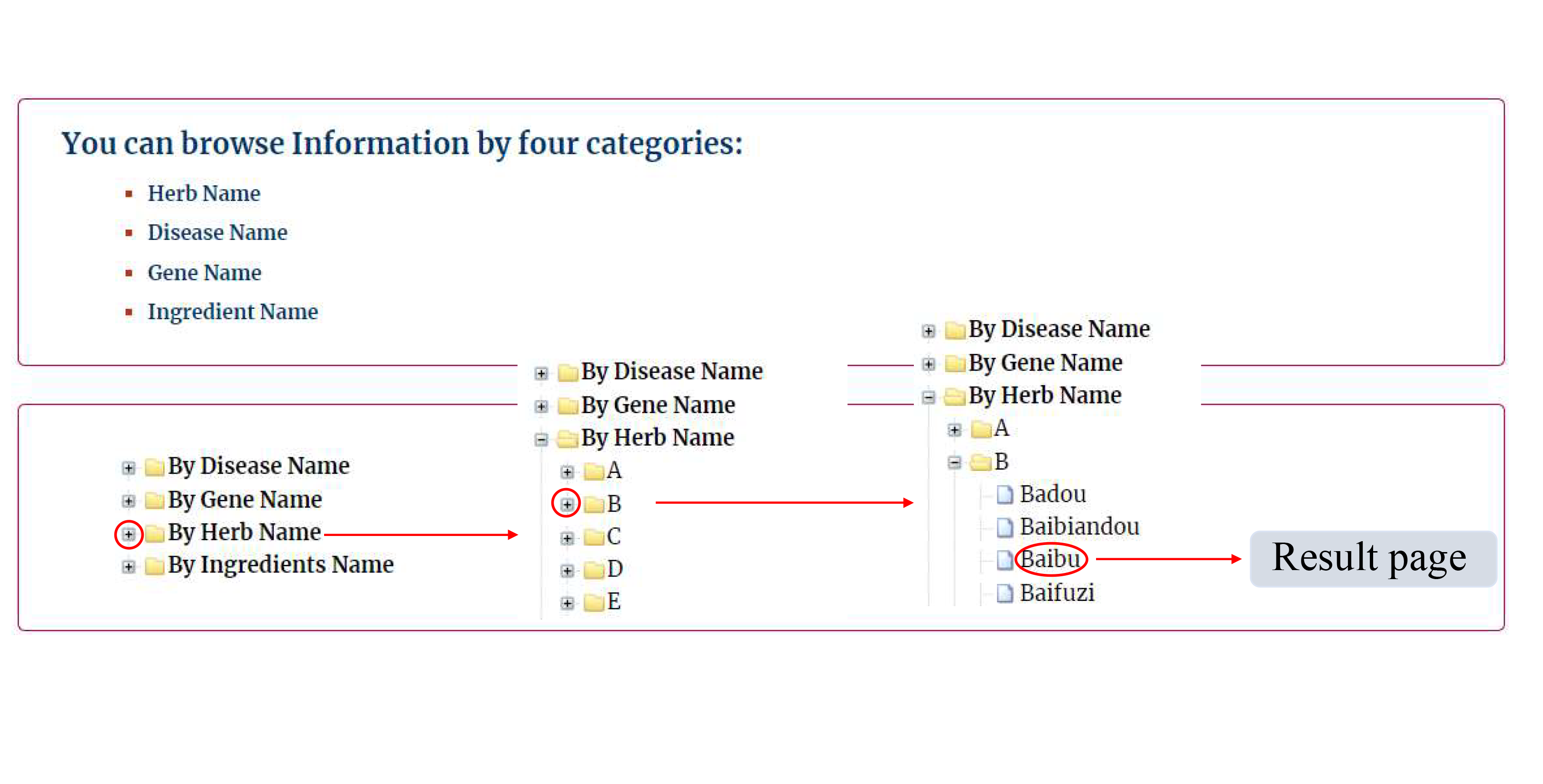
In the Browse page, users can browse the database according to Herb Name, Disease Name, Ingredient Name and Gene Name,respectively. Once clicking the items under each category, the results will be shown in a new page.
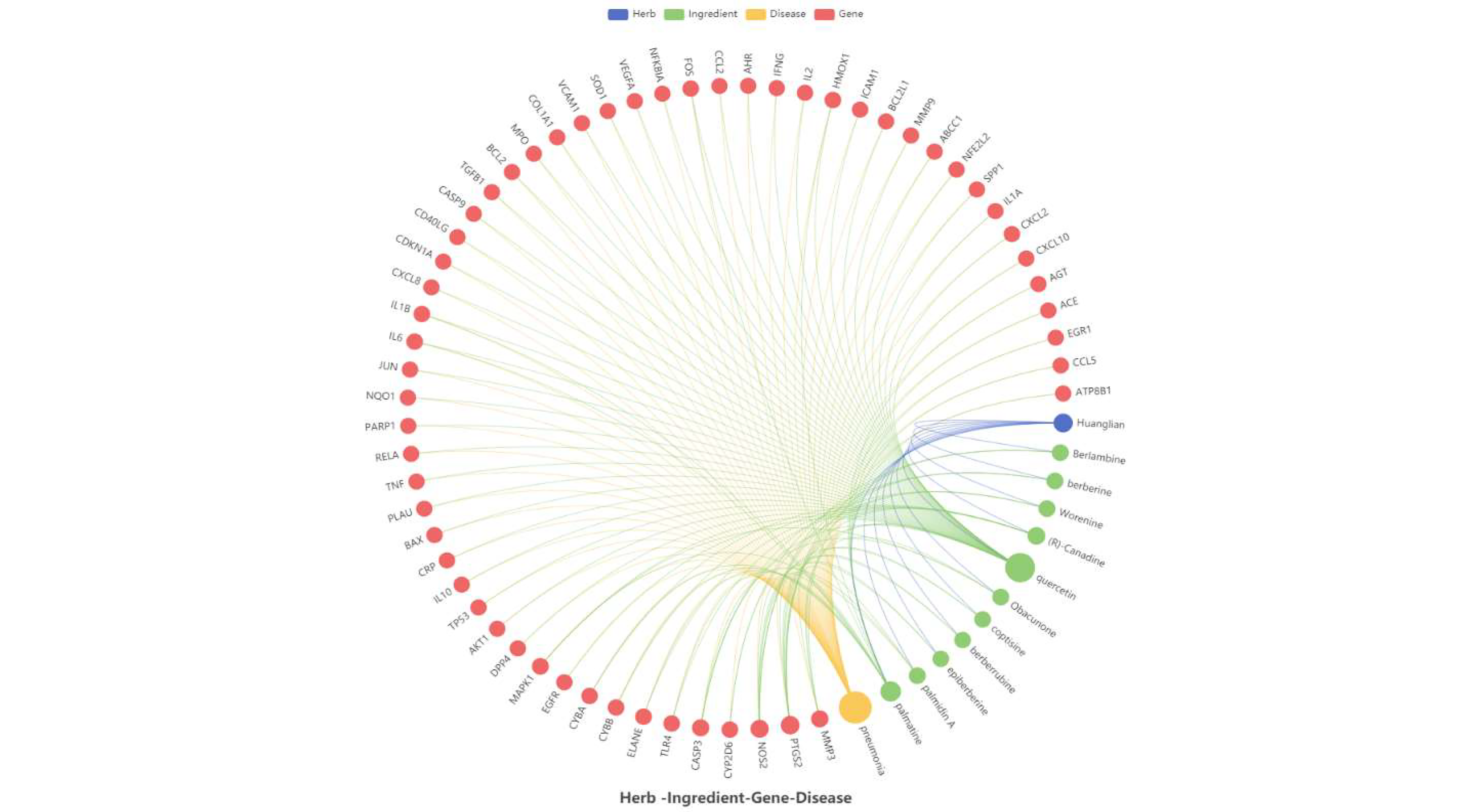
(1) Herb-Ingredient-Gene-Disease Network. Different colored nodes represent different items, The size of each node is related to the connection with other nodes.
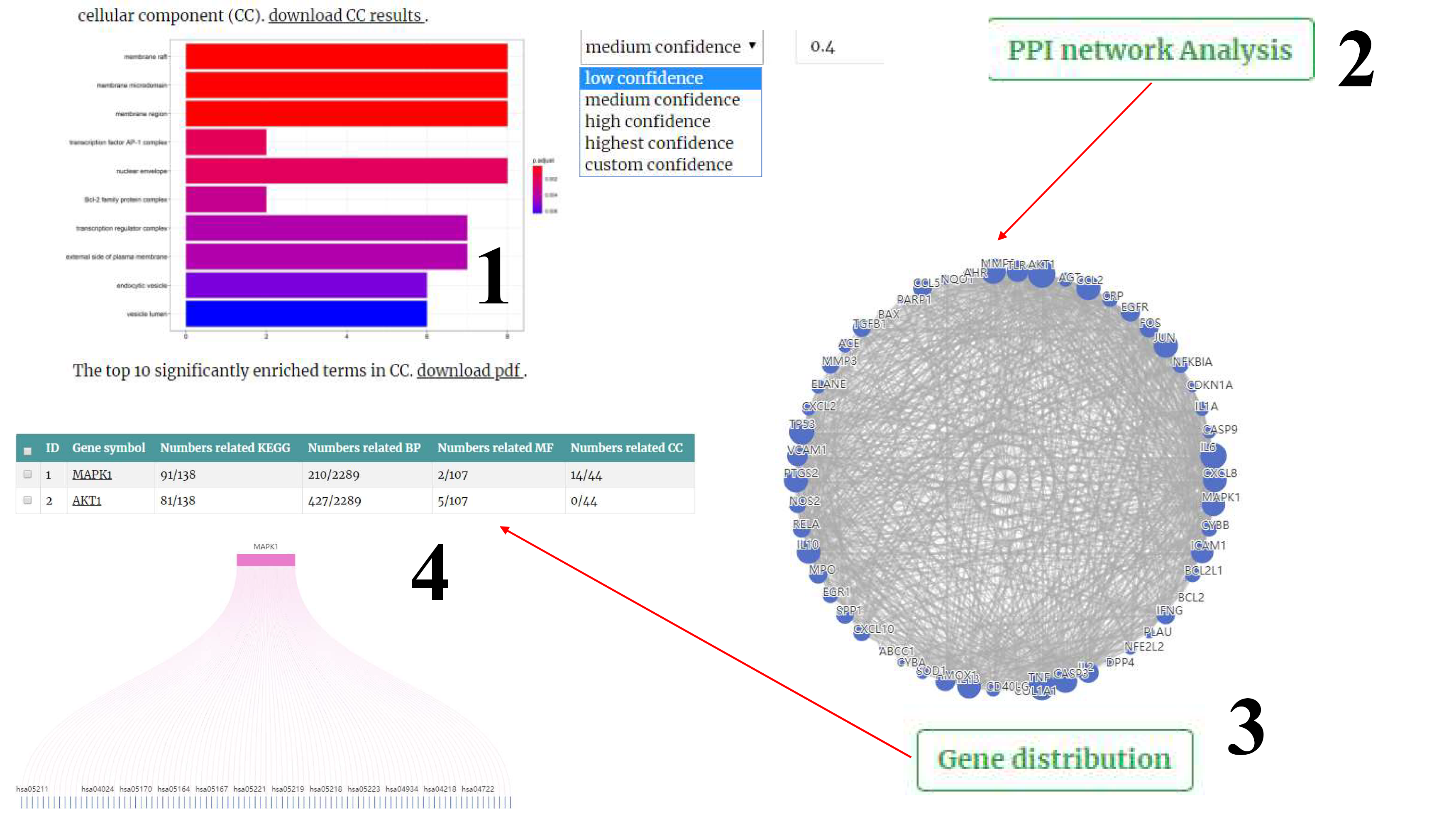
(2) GO and KEGG Pathway Enrichment Analysis. Gene Ontology (GO) annotation is used to define and describe the functions of gene products from three aspects, namely, biological processes (BPs), cell components (CCs), and molecular functions (MFs). The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database that integrates genome, chemistry, and system function information. Users can click the download button, the GO and KEGG analysis results will be obtained.
(3) PPI Network Analysis. Users can customize the confidence score according to their needs, and then clicking the "PPI network Analysis" button, the results will be shown.
(4) Gene distribution. In this page, Users can get the GO and KEGG pathways of gene participation by inputting genes.
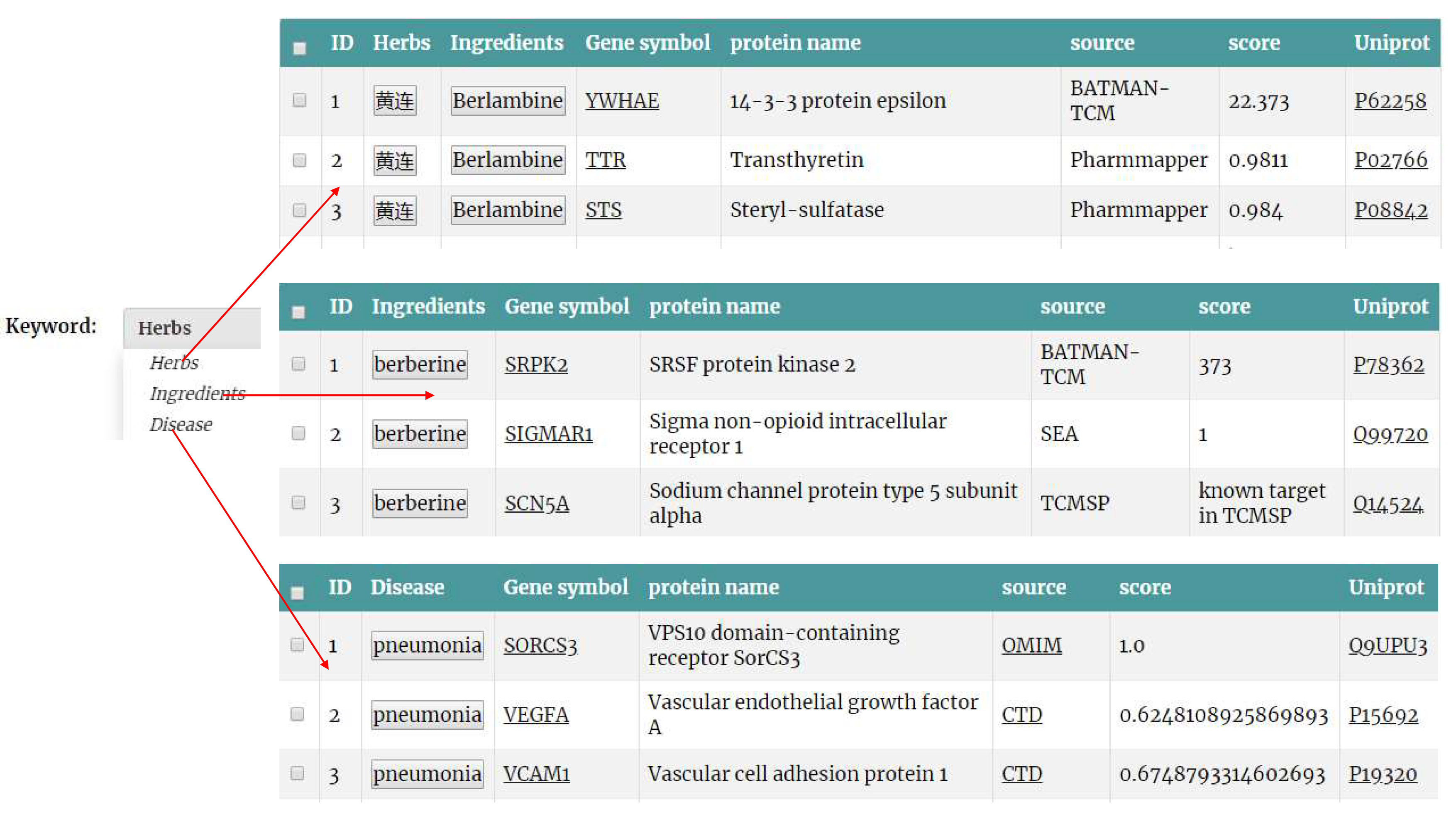
(5) The results of data query. The data query result page shows the results that users are interested in. The provided information includes Herb name, Ingredients name, Gene symbol, protein name, source, score, Uniprot.